|
|

|

|
CABDyN Projects
|
|
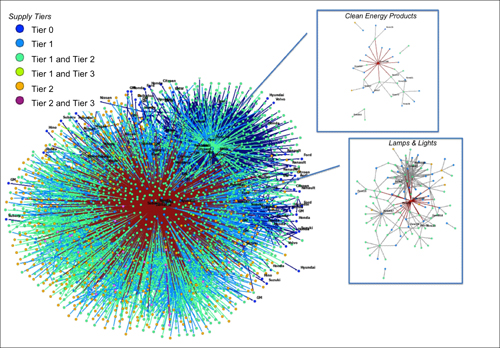
Complex global supply networks
Global supply networks are becoming increasingly complex, with structures that do not correspond to simple linear supply chains or purely hierarchical arrangements, but to date we have lacked large-scale empirical studies that tell us what real-world supply networks look like. This project seeks to address this challenge by focusing on a large, real-world supply network in the automotive industry, with a view to answering questions about structural complexity, dynamics, efficiency, and resilience. The starting point for this research is to construct a large-scale empirical dataset that maps out the relationships between thousands of firms that comprise the Toyota supply network. One immediate finding is that lateral relationships between suppliers in the same tier are not uncommon, so that the real-world structure of the supply network is rather more complex than the default assumption of a pure hierarchy. The analysis of topological network characteristics is augmented and strengthened by a complementary analysis focusing on firm-level properties, such as product portfolios and geographical positioning. Detailed research questions that we would like to address include a longitudinal analysis of how the supply network changes over time, studies of the interdependencies between supply networks of multiple assemblers, and the relationship between network structure and firm performance and strategy. One of the longer-term objectives of the project is to move the field of supply chain management beyond stylised facts, and to demonstrate how a complex networks perspective can contribute novel insights.
Contact: Alexandra Brintrup (CABDyN), Tomomi Kito (BT Centre and CABDyN)
Collaborators: Eduardo López (CABDyN), Steve New, Felix Reed-Tsochas (CABDyN).
|
A Mathematical Model for the Dynamics and Synchronization of Cows
The study of collective behavior of animals, mechanical
systems, or even abstract oscillators has fascinated a large number of
researchers from observational geologists to pure mathematicians. We
consider the collective behavior of herds of cattle. We first consider
some results from an agent-based model and then formulate a mathematical
model for the daily activities of a cow (eating, lying down, and standing)
in terms of a piecewise affine dynamical system. We analyze the
properties of this bovine dynamical system representing the single animal
and develop an exact integrative form as a discrete-time mapping. We then
couple multiple cow "oscillators" together to study synchrony and
cooperation in cattle herds, finding that it is possible for cows to
synchronize less when the coupling is increased.
Reference: arXiv.org (2010)
Contact: Mason A. Porter (CABDyN)
Collaborators: Jie Sun, Erik M. Bollt, Marian S. Dawkins.
|
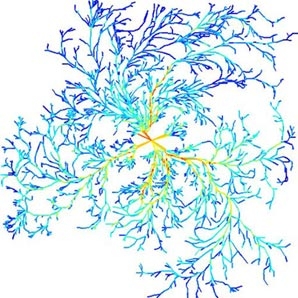
Growth-induced Mass Flows in Fungal Networks
Cord-forming fungi form extensive networks that continuously adapt to maintain an efficient transport system. As osmotically driven water uptake is often distal from the tips, and aqueous fluids are incompressible, we propose that growth induces mass flows across the mycelium, whether or not there are intrahyphal concentration gradients. We imaged the temporal evolution of networks formed by Phanerochaete velutina, and at each stage calculated the unique set of currents that account for the observed changes in cord volume, while minimizing the work required to overcome viscous drag. Predicted speeds were in reasonable agreement with experimental data, and the pressure gradients needed to produce these flows are small. Furthermore, cords that were predicted to carry fast-moving or large currents were significantly more likely to increase in size than cords with slow-moving or small currents. The incompressibility of the fluids within fungi means there is a rapid global response to local fluid movements. Hence velocity of fluid flow is a local signal that conveys quasi-global information about the role of a cord within the mycelium. We suggest that fluid incompressibility and the coupling of growth and mass flow are critical physical features that enable the development of efficient, adaptive biological transport networks.
Reference: Proceedings of The Royal Society B (2010)
Contact: Nick S. Jones (CABDyN) and Eduardo López (CABDyN)
Collaborators: Luke L. M. Heaton, Philip K. Maini (CABDyN), Mark D. Fricker(CABDyN).
|
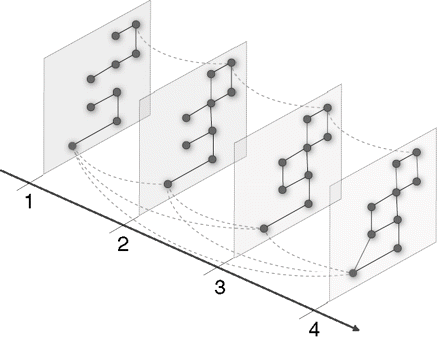
Community Structure in Multislice Networks
One of the most popular and important areas of network science is the study of the "community structure" of a network. A network, usually modelled as a graph, consists of a set of entities and the connections between them. Communities are important because they are thought to have a strong bearing on functional units in many networks. This project entails the investigation of community structure in multislice networks, which can be multiplex (consisting of multiple types of connections), time-dependent, multiscale, and multiplex. We will use recent theoretical advances from our group to examine interesting data sets (which could include, e.g., time series data of H1N1 cases in Mexico) and also generalize the method and consider other ideas (e.g., time-dependent clustering coefficients) on multislice networks.
Reference: Science 328(5980), 876-878 (2010)
Contact: Mason Porter (CABDyN)
Collaborators:
Peter J. Mucha, Thomas Richardson, Kevin Macon, Jukka-Pekka Onnela (CABDyN)
|
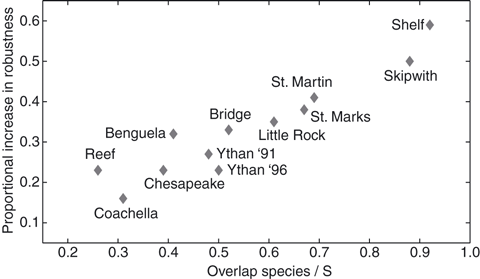
Structural Dynamics and Robustness of Food Webs
Food web structure plays an important role when determining robustness to cascading secondary extinctions. However, existing food web models do not take into account likely changes in trophic interactions ('rewiring') following species loss. We investigated structural dynamics in 12 empirically documented food webs by simulating primary species loss using three realistic removal criteria, and measured robustness in terms of subsequent secondary extinctions. In our model, novel trophic interactions can be established between predators and food items not previously consumed following the loss of competing predator species. By considering the increase in robustness conferred through rewiring, we identify a new category of species – overlap species – which promote robustness as shown by comparing simulations incorporating structural dynamics to those with static topologies. The fraction of overlap species in a food web is highly correlated with this increase in robustness; whereas species richness and connectance are uncorrelated with increased robustness. Our findings underline the importance of compensatory mechanisms that may buffer ecosystems against environmental change, and highlight the likely role of particular species that are expected to facilitate this buffering.
Reference: Ecology Letters (2010)
Contact: Phillip Staniczenko (CABDyN)
Collaborators: Owen T. Lewis, Nick S. Jones (CABDyN), Felix Reed-Tsochas (CABDyN).
|
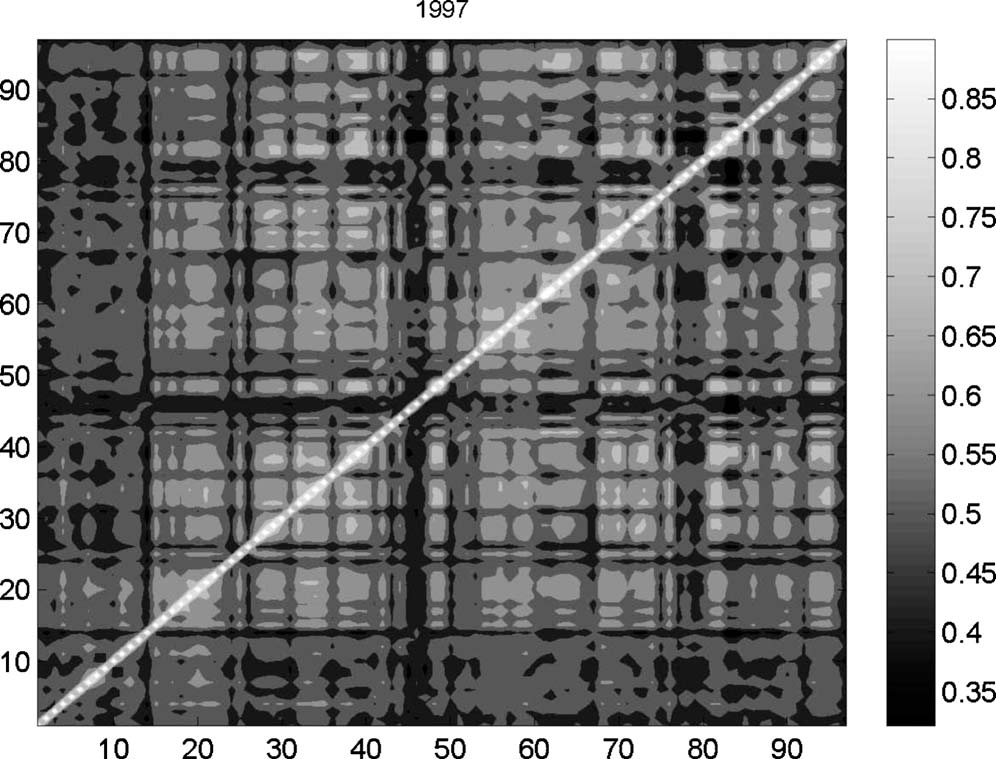
Multinetwork of International Trade: A Commodity-specific Analysis
We study the topological properties of the multinetwork of commodity-specific trade relations among world countries over the 1992–2003 period, comparing them with those of the aggregate-trade network, known in the literature as the international-trade network (ITN). We show that link-weight distributions of commodity-specific networks are extremely heterogeneous and (quasi) log normality of aggregate link-weight distribution is generated as a sheer outcome of aggregation. Commodity-specific networks also display average connectivity, clustering, and centrality levels very different from their aggregate counterpart. We also find that ITN complete connectivity is mainly achieved through the presence of many weak links that keep commodity-specific networks together and that the correlation structure existing between topological statistics within each single network is fairly robust and mimics that of the aggregate network. Finally, we employ cross-commodity correlations between link weights to build hierarchies of commodities. Our results suggest that on the top of a relatively time-invariant “intrinsic” taxonomy (based on inherent between-commodity similarities), the roles played by different commodities in the ITN have become more and more dissimilar, possibly as the result of an increased trade specialization. Our approach is general and can be used to characterize any multinetwork emerging as a nontrivial aggregation of several interdependent layers
Reference: Physical Review E81 (2010)
Contact: Diego Garlaschelli(CABDyN)
Collaborators: Mateo Barigozzi, Giorgio Fagiolo.
|

Rules for Biologically Inspired Adaptive Networks Design
Transport networks are ubiquitous in both social and biological systems. Robust network performance involves a complex trade-off involving cost, transport efficiency, and fault tolerance. Biological networks have been honed by many cycles of evolutionary selection pressure and are likely to yield reasonable solutions to such combinatorial optimization problems. Furthermore, they develop without centralized control and may represent a readily scalable solution for growing networks in general. We show that the slime mold Physarum polycephalum forms networks with comparable efficiency, fault tolerance, and cost to those of real-world infrastructure networks—in this case, the Tokyo rail system. The core mechanisms needed for adaptive network formation can be captured in a biologically inspired mathematical model that may be useful to guide network construction in other domains.
Reference: Science Vol. 327 (2010)
Contact: Mark D. Fricker (CABDyN)
Collaborators: Atsushi Tero, Seiji Takagi, Tetsu Saigusa, Kentaro Ito, Dan P. Bebber (CABDyN).
|
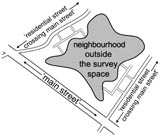
Bias in Epidemiological Studies of Conflict Mortality
Cluster sampling has recently been used to estimate the mortality in various conflicts around the world. A recent study on Iraq employed a new variant of this methodology as far as its microlevel details are concerned by sampling households on "residential streets crossing main streets". This can introduce a substantial bias into the mortality estimates because the residents on residential streets are more likely to be exposed to violence than those living further away. This type of non-coverage bias is notoriously difficult to assess. We apply a modeling approach to gauge the extent of the bias and provide a sensitivity analysis to help readers tune their own judgements on the extent of the bias.
Reference: (2008)
Journal of Peace Research: 45 (5) pp. 653-663
Contact: Jukka-Pekka Onnela (CABDyN)
Collaborators: Neil F. Johnson (CABDyN), Michael Spagat, Sean Gourley, and Gesine Reinert (CABDyN)
|
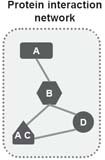
Predicting & Validating Protein Interactions Using Network Structure
Protein interactions play a vital part in the function of a cell. As experimental techniques for detection and validation of protein interactions are time consuming, there is a need for computational methods for this task. Protein interactions appear to form a network with a relatively high degree of local clustering. In this paper we exploit this clustering by suggesting a score based on triplets of observed protein interactions. The score utilises both protein characteristics and network properties. Our score based on triplets is shown to complement existing techniques for predicting protein interactions, outperforming them on data sets which display a high degree of clustering. The predicted interactions score highly against test measures for accuracy. Compared to a similar score derived from pairwise interactions only, the triplet score displays higher sensitivity and specificity. By looking at specific examples, we show how an experimental set of interactions can be enriched and validated. As part of this work we also examine the effect of different prior databases upon the accuracy of prediction and find that the interactions from the same kingdom give better results than from across kingdoms, suggesting that there may be fundamental differences between the networks. These results all emphasize that network structure is important and helps in the accurate prediction of protein interactions. The protein interaction data set and the program used in our analysis, and a list of predictions and validations, are available at http://www.stats.ox.ac.uk/bioinfo/resources/PredictingInteractions.
Reference: (2008)
PLoS Computational Biology: 4 (7) e1000118 (pdf)
Contact: Gesine Reinert (CABDyN)
Collaborators: Pao-Yang Chen and Charlotte M. Deane
|
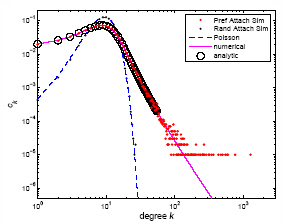
Accelerating Networks
In many real-world, evolving networks, the rate at which links are added to the network is different from the rate at which nodes are added to it, the World WideWeb being one example. This results in the concept of accelerating or decelerating networks. In Accelerating Networks we review existing definitions of network acceleration and, having found them somewhat restrictive, propose a new definition. This might be used to describe a network at any stage of its evolution (whether decaying, growing, or neither), and applies to both directed and undirected networks. We demonstrate its applicability through a simple case study application to the evolution of Wikepedia in three languages.
Existing methods to analyse accelerating networks have used the (somewhat inappropriate) mean-field technique. In ongoing work, we show that it is not only possible, but more accurate to use the master equation technique. This is demonstrated in the graph on the right where the latter is compared to simulated accelerating networks with preferential and random attachment. We demonstrate this process through application to the evolution of the coauthorship network in the cond-mat archive. Preprint will be posted when available.
Reference: New Journal of Physics 9 181
Contact: David Smith (CABDyN)
Collaborators: Neil F. Johnson (CABDyN) and Jukka-Pekka Onnela (CABDyN)
|
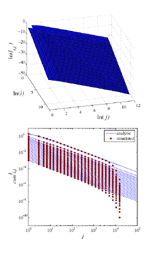
Link-space formalism for network analysis
We introduce the link-space formalism for analyzing network models with degree-degree correlations. The
formalism is based on a statistical description of the fraction of links li,j connecting nodes of degrees i and j.
To demonstrate its use, we apply the framework to some pedagogical network models, namely, random
attachment, Barabási-Albert preferential attachment, and the classical Erdos and Rényi random graph. For
these three models the link-space matrix can be solved analytically. We apply the formalism to a simple
one-parameter growing network model whose numerical solution exemplifies the effect of degree-degree
correlations for the resulting degree distribution. We also employ the formalism to derive the degree distributions
of two very simple network decay models, more specifically, that of random link deletion and random
node deletion. The formalism allows detailed analysis of the correlations within networks and we also employ
it to derive the form of a perfectly nonassortative network for arbitrary degree distribution.
Reference:
Physical Review E. 77 036112. [pdf]
Contact: David Smith (CABDyN)
Collaborators: Chiu Fan Lee(CABDyN), Jukka-Pekka Onnela (CABDyN), and Neil F Johnson (CABDyN).
|
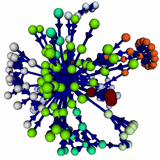
 Structure and tie strengths in mobile communication networks Structure and tie strengths in mobile communication networks
Removal of weak links from a mobile communication network sample (frame 1) causes a phase transition from a global network to a collection of unconnected "islands" (frame 2), whereas global connectivity is maintained if strong ties are removed (frame 3). These findings support the qualitatively different global role played by weak and strong ties in large-scale social networks.
Reference: Adapted from 'Structure and tie strengths in mobile communication networks'.
Proceedings of the National Academy of Sciences U.S.A. (PNAS) 104, 7332-7336 (2007).
Contact: Jukka-Pekka Onnela (CABDyN)
Collaborators: J. Saramäki, J. Hyvönen, G. Szabó, D. Lazer, K. Kaski, J. Kertész, and A.-L. Barabási
|
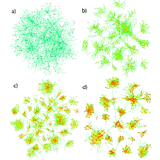
Emergence of Communities in Weighted Networks
Topology and weights are closely related in complex networks and this is reflected in their modular structure. We introduce a new network model in which the weights are generated dynamically and shape the developing topology. By tuning a model parameter that governs the model's sensitivity to weights, the resulting networks undergo a gradual structural transition from a module-free topology (a) to one with clear communities (d). As the underlying microscopic mechanisms are sociologically plausible, the model can be used to explain the emergence of communities in social networks.
Reference: Physical Review Letters. 99, 228701.
Contact: Jukka-Pekka Onnela (CABDyN)
Collaborators: Jussi M. Kumpula, Jari Saramäki, Kimmo Kaski, and János Kertész.
|

Mechanisms for robust network contraction
This project seeks to complement the large body of work on network growth by developing new models which explore how networks may contract without collapsing, and what specific mechanisms can be identified that contribute to the robustness of a contracting network. We therefore need to understand how different combinations of assembly and disassembly processes act on network structures, and under what circumstances the network topology is preserved. The empirical side of the project is based on an extensive dataset of transactions between firms in the New York garment industry over a period of almost 20 years. In addition to the empirical research, extensive simulations are run to explore different modes of contraction, and to generalize the findings beyond the specific inter-firm network on which the empirical research is based.
Reference: 'Asymmetric disassembly and robustness in declining networks',
Proceedings of the National Academy of Sciences U.S.A . (PNAS) 105, 16466-16471 (2008).
Contact: Felix Reed-Tsochas (CABDyN)
Collaborators: Serguei Saavedra (CABDyN) and Brian Uzzi
|
Interaction patterns in ecological and organizational networks
This project looks at empirical data for a specific class of ecological networks, as well as data from a manufacturer-contractor network, with a view to identifying common features in the interaction patterns for these very different systems and environmental contexts. The similarities that we find are not simply fortuitous, but reflect that in both cases there are analogous constraints on consumer-resource interactions. The empirical analysis is augmented by the aim to develop a stochastic model that can account for the observed structural features in both types of networks.
Reference: 'A simple model of bipartite cooperation for ecological and organizational networks',
Nature 457, 463-466.
(Advance Online Publication: 3rd December 2008, doi:10.1038/nature07532)
Contact: Felix Reed-Tsochas (CABDyN)
Collaborators: Serguei Saavedra (CABDyN) and Brian Uzzi
|

Transmission of errors in production networks
Production networks emerge from multiple interactions between the participants involved in making a specific item or element. These interactions play a key role in inhibiting or favoring the transmission of information and resources throughout the network, which plays a central role in the correct operation of the system as a whole. In this project we analyze a large dataset of firms involved in a distributed process of production, and use analytic methods and simulations to study the transmission of production errors in organizational networks.
Contact: Felix Reed-Tsochas (CABDyN)
Collaborators: Serguei Saavedra (CABDyN), Janet Smart (CABDyN) and Brian Uzzi
|
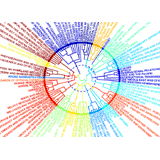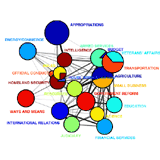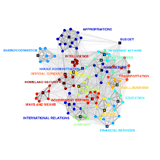 Community Structure in the U.S. House of Respresentatives Community Structure in the U.S. House of Respresentatives
Much of the work involved in making United States law is performed by Congressional committees and subcommittees before bills reach the floors of the Senate and House of Representatives. Committee memberships can be viewed as a social network, and the relationships between legislators appointed to common committees can be grouped into various hierarchical levels: individuals, subcommittees, standing (and select) committees, groups of committees, and the entire floor. Community detection methods from network theory have been successful not only at replicating this hierarchical organization but also at uncovering additional structural features of committee networks. One example, illustrated here for the 108th House (2003--2004), is the strong overlap between certain committees, such as between the Rules Committee and the Select Committee on Homeland Security. (Abstract pdf)
Reference: Physica A 386 414-438 (2007). (pdf)
Contact: Mason Porter (CABDyN)
Collaborators: Peter J. Mucha, M.E.J Newman and A.J. Friend
|
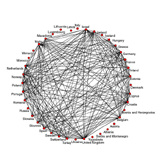
Musical Taste and Voting Networks
How do actors or agents make up their mind? Do they follow their own criteria, or are their choices biased by the behavior of others? Is it common for two or more groups to form which oppose each other? What answers does a study of the Eurovision song contest provide?
References:
Physica A 360, 576-598 (2006)
Physica A 377, 672-688 (2006)
Contact: Janet Smart (CABDyN)
Collaborators: Neil Johnson (CABDyN), Felix Reed-Tsochas (CABDyN) and Serguei Saavedra (CABDyN)
|
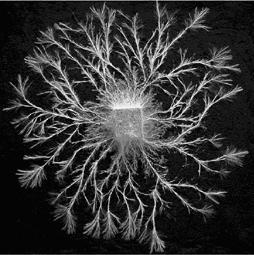
Foraging fungal mycelia as model experimental networks
Many relatively simple organisms, such as bacteria, cellular and acellular slime moulds and fungi, can self-organize to form complex developmental networks with a rich variety of structure and behaviour. Many of these systems are intimately associated with nutrient acquisition or distribution, particularly under conditions where resources are limited and distributed patchily in time and/or space. It is postulated that emergent structures are likely to be efficient transport systems and resilient to attack as they have been subject to many cycles of evolutionary selection pressure. In comparison to many other biological networks, such as neural networks, genetic and biochemical pathways or food webs, microbes are also extremely accessible, and provide tractable experimental systems.
Reference: Bebber et al (2007)'Biological solutions to transport network design'.
Proceedings of the Royal Society B 274 2307-2315. doi:10.1098/rspb.2007.0459. (pdf)
Contact: Mark Fricker (CABDyN)
Collaborators: Dan Bebber (CABDyN), J. Hynes, P.R. Darrah and L. Boddy
|
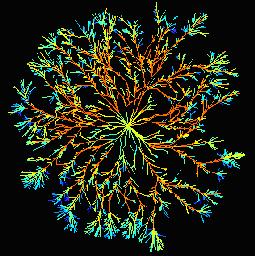 Resilience in planar spatial networks Resilience in planar spatial networks
For example, foraging saprotrophic woodland fungi form extensive interconnected mycelial networks that scavenge for scarce, patchy resources in the leaf litter layer and uppermost soil horizons. The architecture of the network continuously adapts in response to local nutritional or environmental cues, damage or predation, through a combination of growth, branching, fusion (anastomosis) or regression. In collaboration with Prof Lynne Boddy in Cardiff, we have started to characterize the colony topology as a planar spatial network to evaluate foraging efficiency, resilience to attack and overall cost, using graph theory and network analysis.
References
Fricker, et al. (2007) Mycelial networks: structure and dynamics
In: Ecology of Saprotrophic Basiodiomycetes. Eds L. Boddy, J.C. Franklin and P. van West. (in press)
Fricker et al. (2007) Network organisation of mycelial fungi.
In: The Mycota. Vol VIII, Biology of the Fungal Cell (2nd Ed). Eds R.J.Howard and N.A.R. Gow. (in press)
Contact: Mark Fricker (CABDyN)
Collaborators: Dan Bebber (CABDyN), P.R. Darrah, J. Hynes, L. Boddy
|
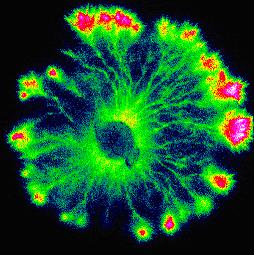 Self-organization of transport dynamics Self-organization of transport dynamics
Embedded within the physical structure is a set of physiological processes that contribute to uptake, storage and redistribution of nutrients throughout the network. We have developed a novel scintillation imaging technique to map solute transport dynamics in fungal mycelial networks. This has revealed a marked pulsatile component to flux superimposed on a rapid underlying translocation. The network seems to self-organize into well demarcated domains that differ in phase of the oscillations. There is also abrupt switching between different pre-existing transport routes. To analyze these complex events we are developing Fourier, Hilbert and wavelet based phase-mapping techniques.
References:
Tlalka, et al. (2003) New Phytol. 158:325-335,
Tlalka, et al. (2002) New Phytol. 153:173-184.
Fricker et al. (2007) Fung. Genet. Biol. (in press)
Tlalka et al. (2007) Fung. Genet. Biol. (in press)
Bebber et al. Imaging complex nutrient dynamics in mycelial networks.
In: G.M. Gadd (Ed). Fungi in the Environment. Cambridge University Press, Cambridge. (in press)
Contact: Mark Fricker (CABDyN)
Collaborators: Dan Bebber (CABDyN), M. Tlalka, J. Hynes, P.R. Darrah, A. Ashford,
S.C. Watkinson, L. Boddy, D Hensman and S. Takagi.
|
 Agent-based modeling of resource canalisation Agent-based modeling of resource canalisation
Mark Fricker, Neil Johnson, David Smith and Chiu-Fan Lee have developed a generic agent-based model which is remarkably simple, yet which yields emergent structural networks that capture the dynamical evolution of canalised nutrient fluxes observed in real fungi. In this model, the nutrients required for growth are translocated through the developing structure by the agents themselves. This is in contrast to agent models based on diffusion-limited aggregation (DLA) or diffusion-limited growth (DLG), and most continuous (PDE) models where growth is controlled by diffusion of nutrients through an external field.
Reference:
Smith et al. (2008) (pdf)
Contact: Mark Fricker (CABDyN)
Collaborators: David Smith (CABDyN), Neil Johnson (CABDyN) ,
Chiu-Fan Lee (CABDyN), Jukka-Pekka Onnela (CABDyN)
|
 3D branching fungal mycelia 3D branching fungal mycelia
We are also collaborating with Prof. David Moore and Audrius Meskauskas at Manchester to explore the use of their Neighbour-Sensing mathematical model of hyphal growth to generate 3-D branching trees. The Neighbour-Sensing model brings together the basic essentials of hyphal growth into a vector-based mathematical model. The rate of hyphal growth and branching is incluenced by the surrounding virtual mycelium, and external factors. The program is implemented in Java to run on stand-alone systems and also supercomputer clusters.
Meškauskas et al. (2004) Mycol. Res. 108:1-16.
See: http://www.world-of-fungi.org/
Contact: Mark Fricker (CABDyN)
Collaborators: D. Moore and A. Meškauskas
|
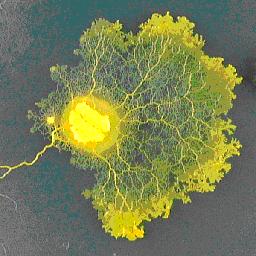 Network organization in slime molds Network organization in slime molds
A series of exchange visits have started a new collaboration with Toshiyuki Nakagaki in the Creative Research Institute at the University of Hokkaido, Japan. This is to investigate how the network organization of the infamous Physarum slime mold adapts to particular geometric configurations of resources, nutrient gradients and also imposed stress gradients. Neil Johnson and his colleagues have already developed analytical mathematical models for some of these network situations, whilst Toshiyuki Nakagaki and his colleagues have developed powerful biologically-inspired models of network formation.
Contact: Mark Fricker (CABDyN)
Collaborators: Neil Johnson (CABDyN) and T. Nakagaki
|
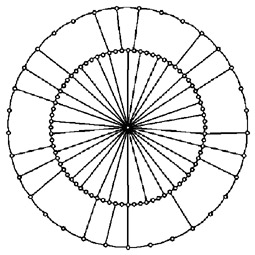 Abstracted network models inspired by fungi Abstracted network models inspired by fungi
An alternative approach to understand the behaviour of fungal networks is to abstract the essence of a mycelial network and recast it in a form that permits more rigorous mathematical analysis. Thus the interplay between radial expansion and lateral connections can be captured in a ‘hub-and-spoke’ model of the developing fungal mycelium (Ashton et al., 2005; Jarrett et al., 2006) that allows exactly solvable solutions to questions of nutrient flows under different cost scenarios. The the average shortest path length across the network exhibits non-trivial behaviour when different cost functions are imposed on transport through the hub, for example. It is possible to extend this model further to consider a more complicated model of a biological system which contain a ring and hub embedded within another ring, with the original ring-hub motif functioning now as a hub by renormalizing each ring-hub combination.
Reference: Jarrett et al. (2006) Physical Review E 74 026116
Contact: Mark Fricker (CABDyN)
Collaborators: T. Jarrett, D. Ashton and Neil Johnson (CABDyN)
|
|
|

|

|